Showing posts with label fragment growth. Show all posts
Showing posts with label fragment growth. Show all posts
Tuesday, June 30, 2020
Tutorials of 3D drug design by AI and de novo method
1. Introduction
In this tutorial, the standard protocol of MolAICal (https://doi.org/10.1093/bib/bbaa161) is introduced for the drug design of glucagon receptor (GCGR) by artificial intelligence and de novo method. It will help the pharmacologist, chemists and other scientists design rational drugs according to three-dimensional active pocket of proteins.
2. Materials
2.1. Software requirement
1) MolAICal: https://molaical.github.io
2) UCSF Chimera: https://www.cgl.ucsf.edu/chimera/
2.2. Protocol files
All the necessary tutorial files are downloaded from:
https://github.com/MolAICal/tutorials/tree/master/001-AIGrow
For more detailed procedures, please go to https://molaical.github.io, and click the section named "Tutorials" in the left part of webpage. Once "Tutorials" open, you can freely find pdf file about this tutorial (see Figure 1):
Figure 1
You can repeat this tutorial according to the content of pdf file.
Quick start for de novo drug design of COVID-19 Mpro by MolAICal
Introduction
SARS-CoV-2 caused the rapid spread of coronavirus disease 2019 (COVID-19) throughout the world. In this tutorial, the SARS-CoV-2 main protease (Mpro) which plays an important role on the replication of coronavirus is selected as the example target. The crystal structures of SARS-CoV-2 Mpro have been reported (PDB ID: 6LU7, 6Y2F, etc) [1,2]. Here, the built structure of SARS-CoV-2 Mpro provided by the group of Prof. Zihe Rao is selected as our quick start example (see Figure 1). Here, MolAICal (https://doi.org/10.1093/bib/bbaa161) is employed for this tutorial.
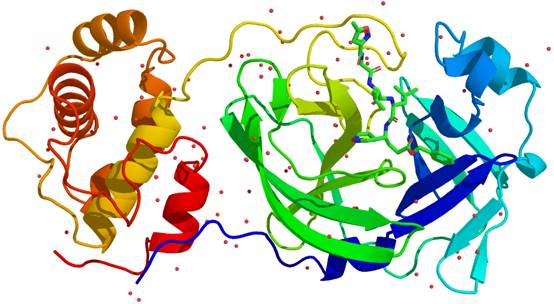
Figure 1. The structure of SARS-CoV-2 Mpro

Figure 1. The structure of SARS-CoV-2 Mpro
Materials
1. Software requirement
1) MolAICal: https://molaical.github.io
2) UCSF Chimera: https://www.cgl.ucsf.edu/chimera/
Make sure every software is installed rightly.
1) MolAICal: https://molaical.github.io
2) UCSF Chimera: https://www.cgl.ucsf.edu/chimera/
Make sure every software is installed rightly.
2. Example files
All the necessary tutorial files are downloaded from: QuickStart
All the necessary tutorial files are downloaded from: QuickStart
Procedure
1. Open “InputParFile.dat” file, it needs to modify four parameters for this tutorial:
------------------------------------------------------------------------------------------
receptorPDB mproNolig.pdb
startFragFile startFrag.mol2
centerPoints -10.733 12.416 68.829
boxLengthXYZ 30.0 30.0 30.0
-------------------------------------------------------------------------------------------
The “receptorPDB” is set to the PDB format structure of SARS-CoV-2 Mpro without ligand. The “startFragFile” is set to the initial fragment. The “centerPoints” and “boxLengthXYZ” represent the box center and length, respectively.
------------------------------------------------------------------------------------------
receptorPDB mproNolig.pdb
startFragFile startFrag.mol2
centerPoints -10.733 12.416 68.829
boxLengthXYZ 30.0 30.0 30.0
-------------------------------------------------------------------------------------------
The “receptorPDB” is set to the PDB format structure of SARS-CoV-2 Mpro without ligand. The “startFragFile” is set to the initial fragment. The “centerPoints” and “boxLengthXYZ” represent the box center and length, respectively.
2. Download and open tutorial folder “000-quickStart”, then run command as below:
#> molaical.exe -denovo grow -i InputParFile.dat
Results
The results are stored in the folder named “001-AIGrow/results”. The “AstatisticsFile.dat” records the information of designed ligands that contain ID, Name, Cluster, Affinity, Formula, InChIKey. The results are just a simple demo example that does not contain complete running results. The complete results can be obtained by finishing this task.
Open crystal ligand N3 named “ligand.mol2” of SARS-CoV-2 Mpro and generated ligand named “lig_11.mol2” (see Figure 2):
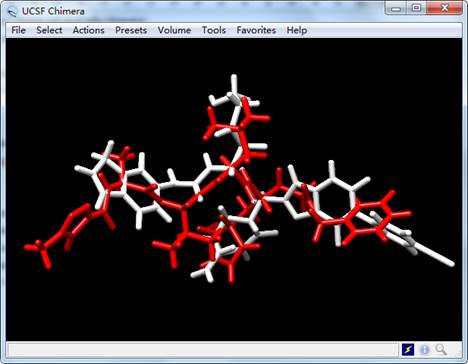
Figure 2. The results of generated ligand (white) and inhibitor N3 (red) of SARS-CoV-2 Mpro.
Open crystal ligand N3 named “ligand.mol2” of SARS-CoV-2 Mpro and generated ligand named “lig_11.mol2” (see Figure 2):

Figure 2. The results of generated ligand (white) and inhibitor N3 (red) of SARS-CoV-2 Mpro.
The detailed tutorial can be referred in the first tutorial of “1. Drug design tutorials of MolAICal” in https://molaical.github.io/
References
[1] Jin, Z. et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature (2020).
[2] Zhang, L. et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science (2020).
[2] Zhang, L. et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science (2020).
Subscribe to:
Posts (Atom)
